KEGG2Net generates a network involving only the genes represented on a KEGG pathway with all of the direct and indirect gene-gene interactions deduced from the pathway. KEGG2Net offers four different methods to remove cycles from the resulting gene interaction networks to construct directed acyclic graphs (DAGs). Click the link below to download the gene interation networks and DAGs for your organism of interest.
Please cite: Chanumolu SK, Albahrani M, Can H, Otu HH. "KEGG2Net: deducing gene interaction networks and acyclic graphs from KEGG pathways" EMBnet.journal, 2021,26:e949. DOI: 10.14806/ej.26.0.949
 | 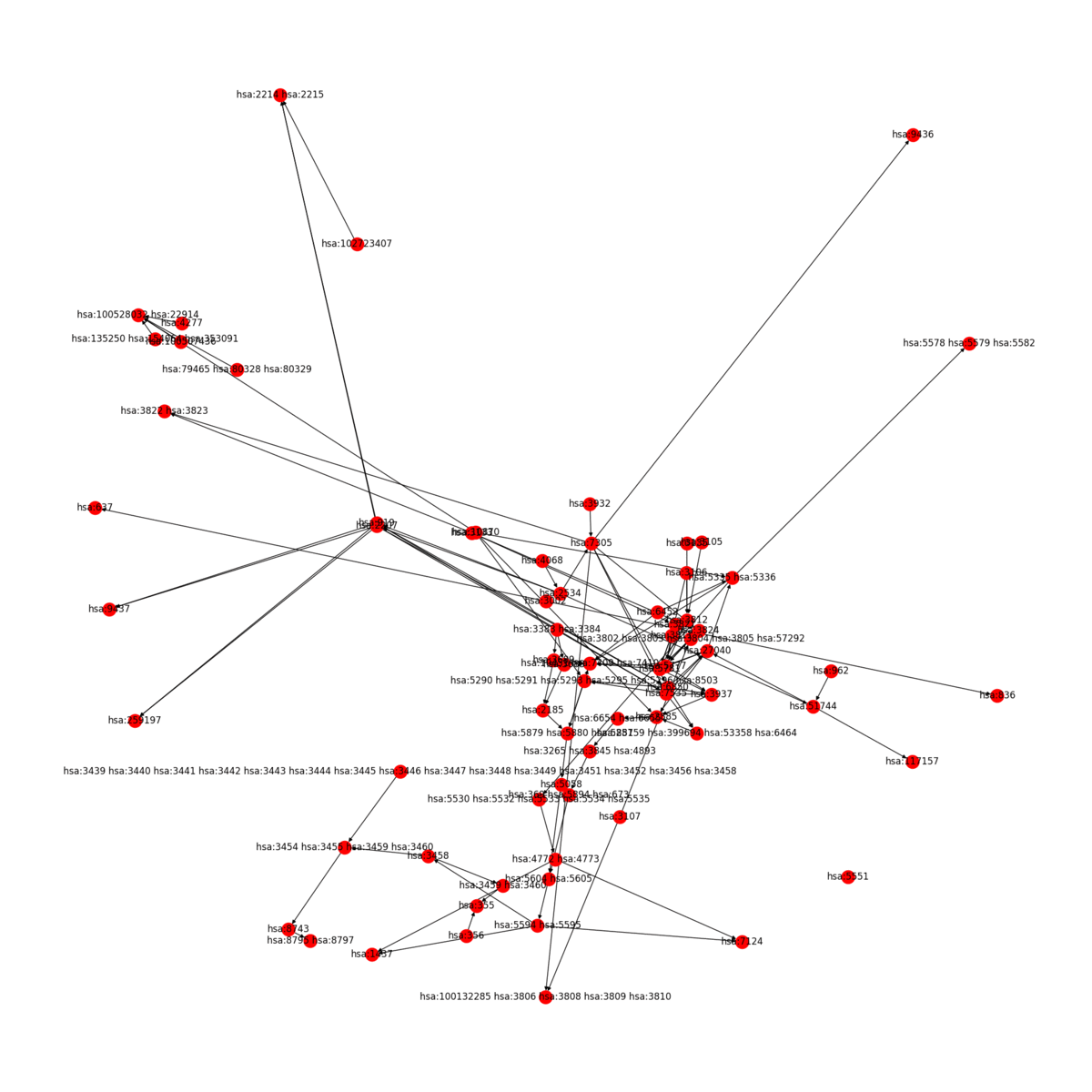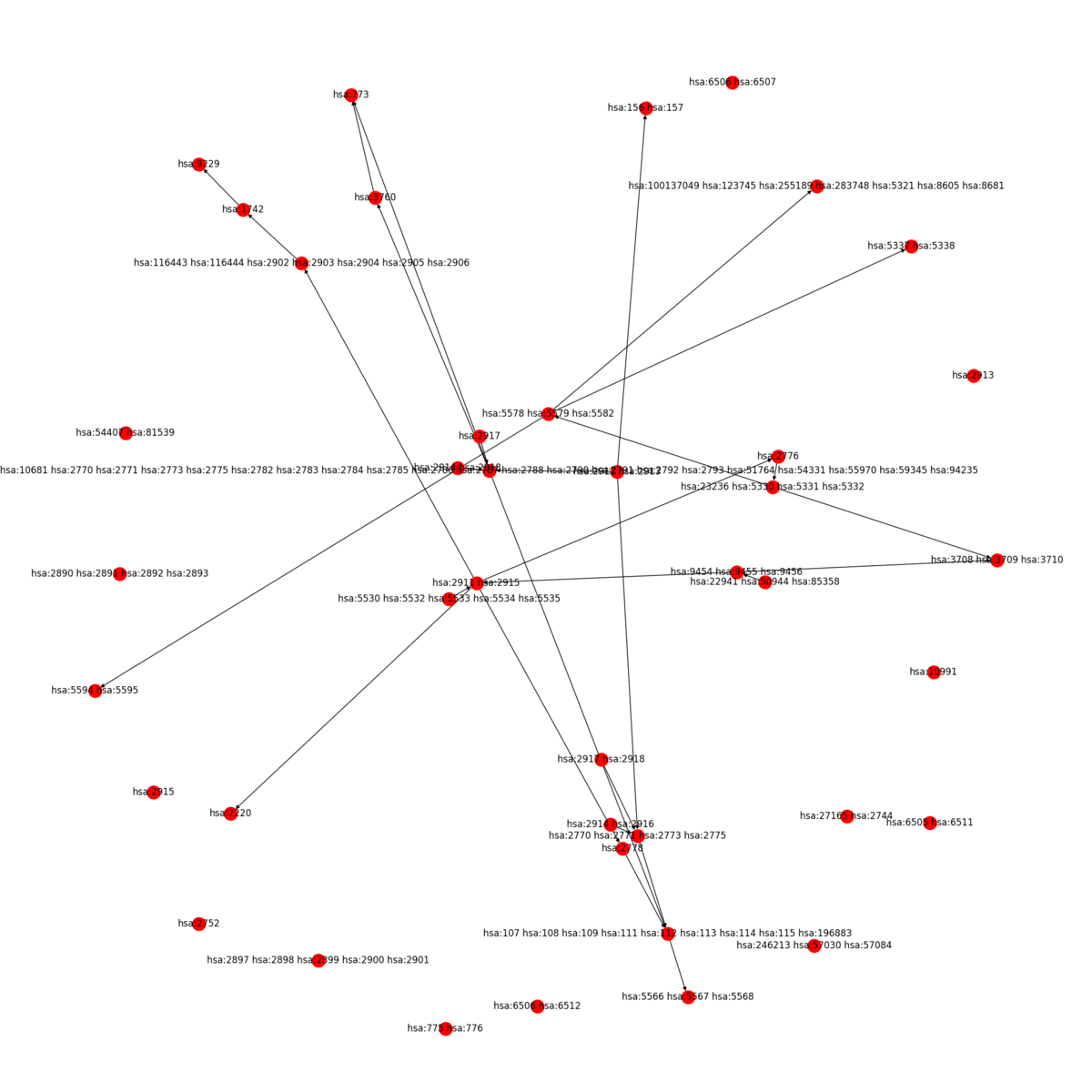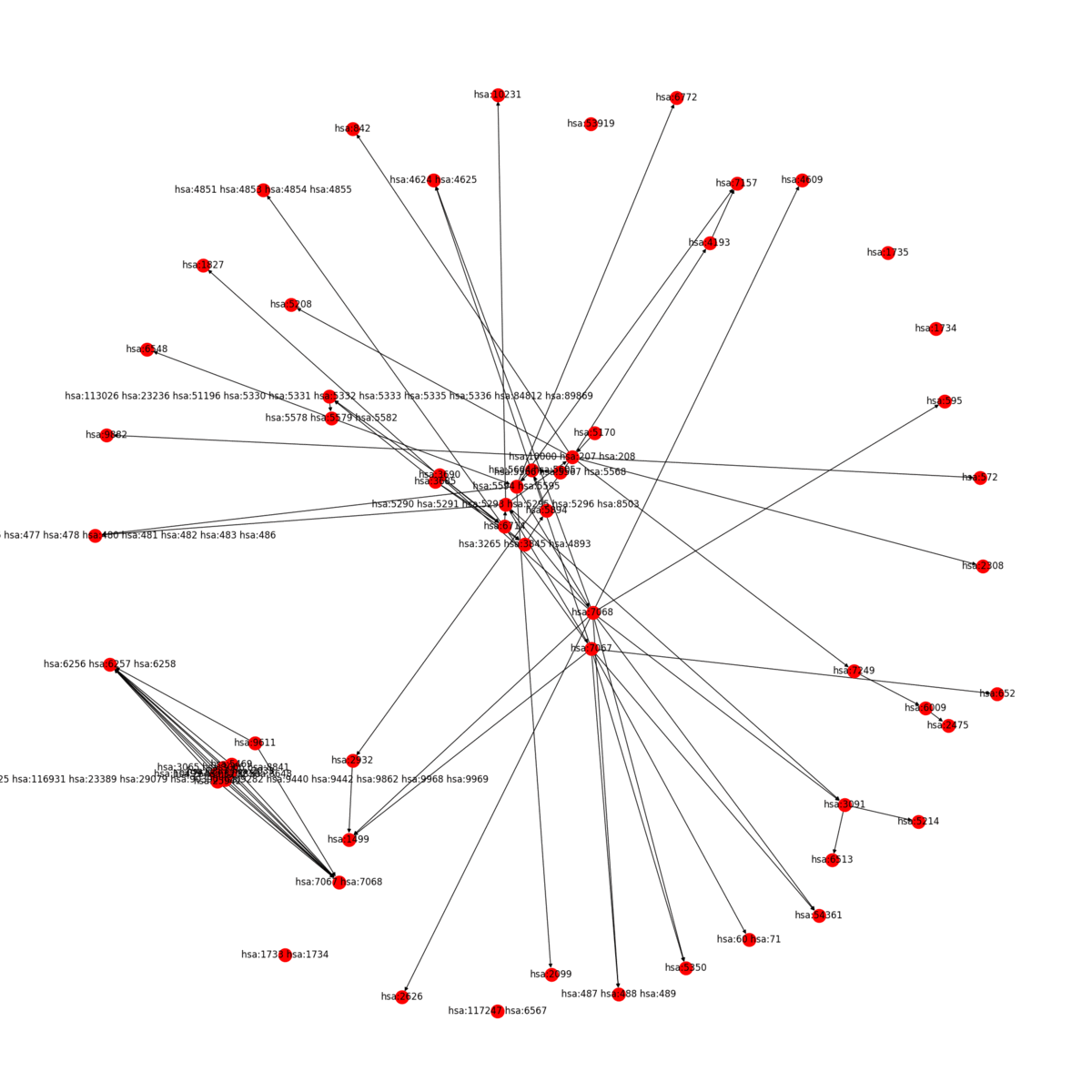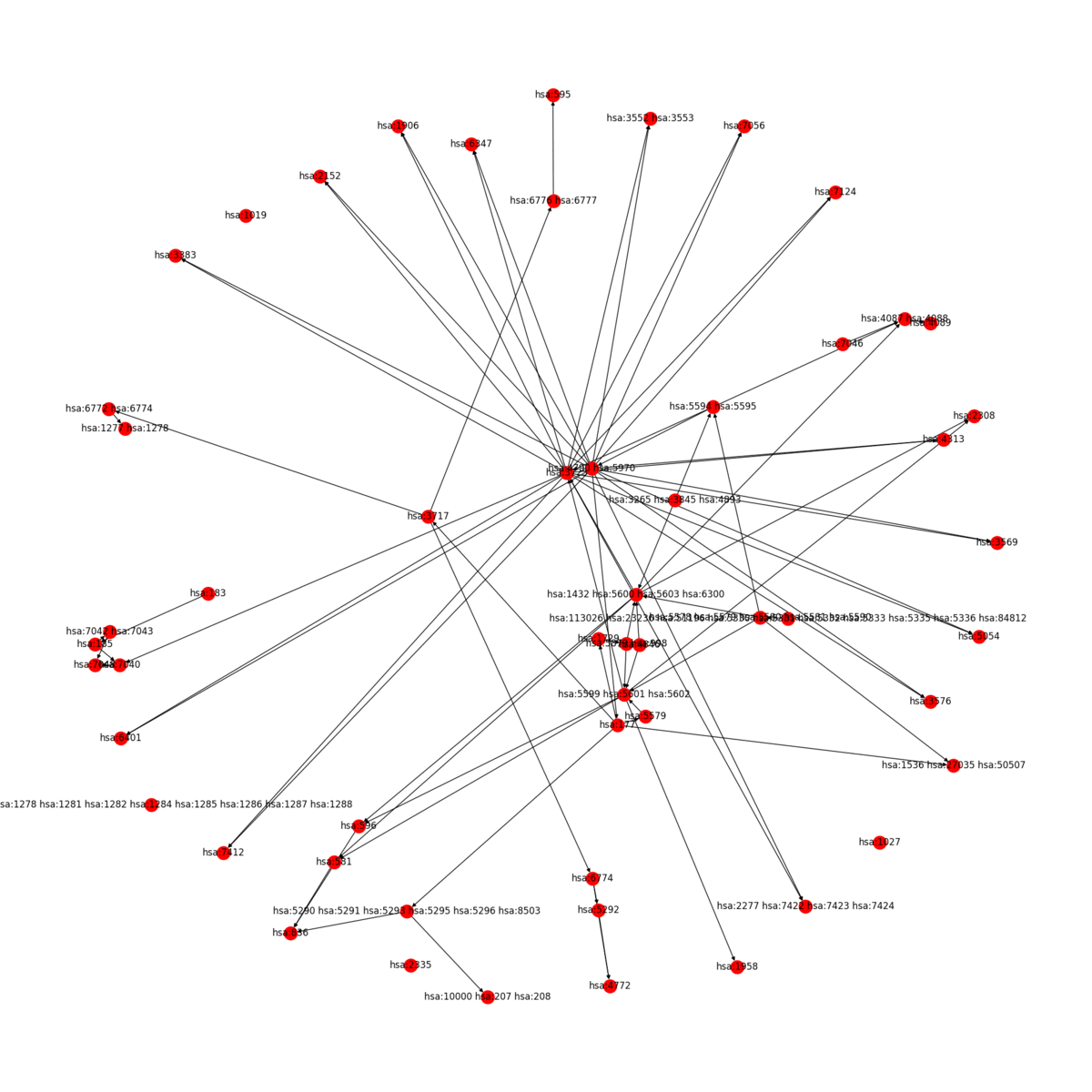 |
KEGG2Net can be used for any organism hosted in KEGG. Sample output for human is given below:
KEGG Pathway IDs and Names
Adjacency matrices (Gene Interaction Networks)
SIF Files (Gene Interaction Networks)
Graphs (Gene Interaction Networks)
Adjacency matrices (DAGs generated with 4 alternative methods)
SIF Files (DAGs generated with 4 alternative methods)
Graphs (DAGs generated with 4 alternative methods)
Contact Us:
Hasan Otu, Ph.D.
Email: hotu2@unl.edu
Handan Can, Ph.D.
Email: handancan03@gmail.com
Sree Krishna Chanumolu, Ph.D.
Email: instanceofsree@gmail.com
Electrical and Computer Engineering, University of Nebraska–Lincoln, 430F NH, Lincoln, NE 68588-0511
Visit OTULAB
Development of KEGG2Net was supported by the National Library of Medicine (NLM) of the National Institutes of Health (NIH) under award number R21LM012759.