Software
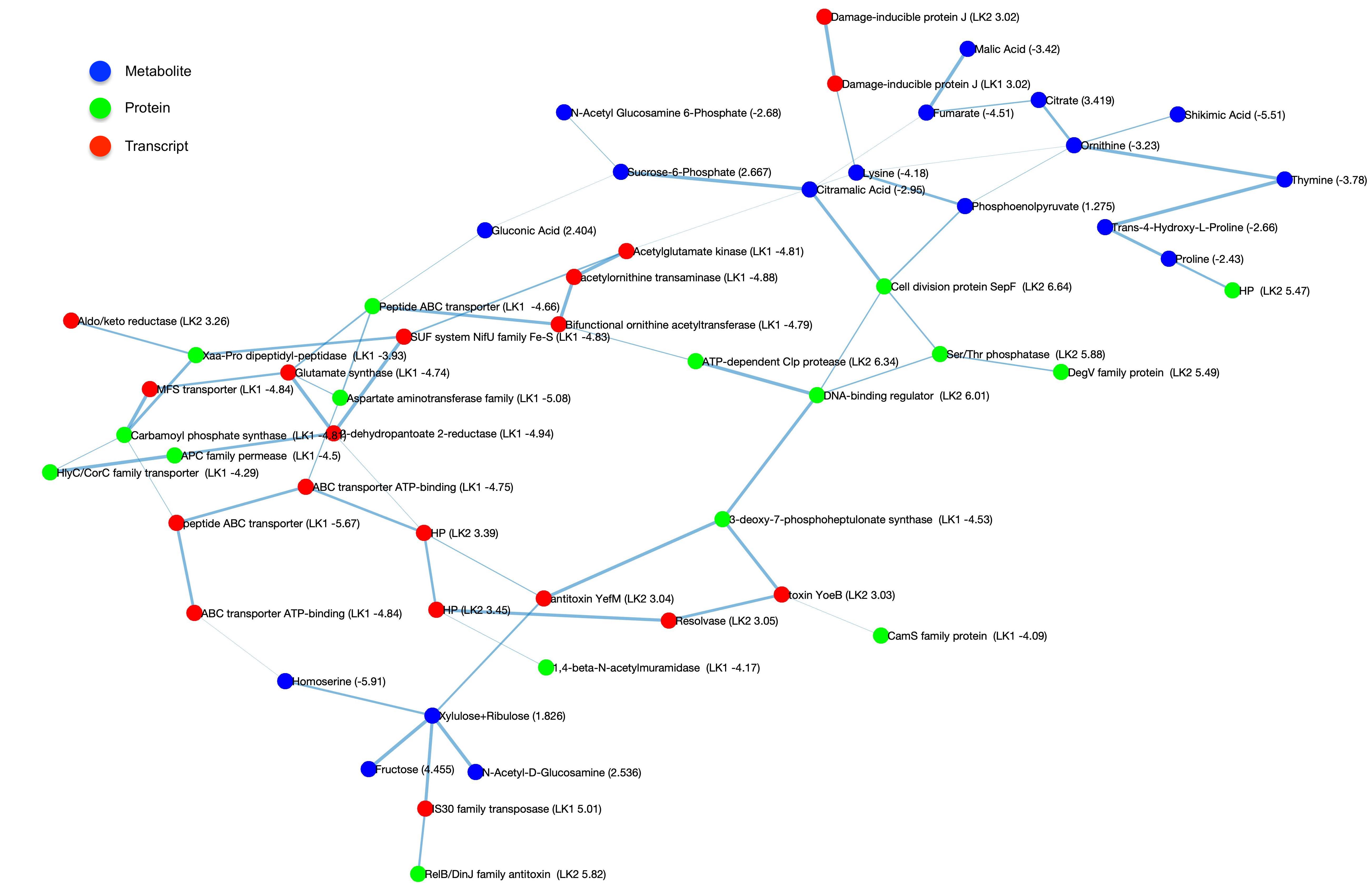
MEMINEX MEta-Multi-omics Integration using Networks and EXternal knowledge builds interaction networks for multiomics data from a microbial community using probabilistic graph representations and external knowledge. MEMINEX indicates the organisms from which the interacting molecules come from and succesfully discovers multifarious interactions between different omics types. Citation
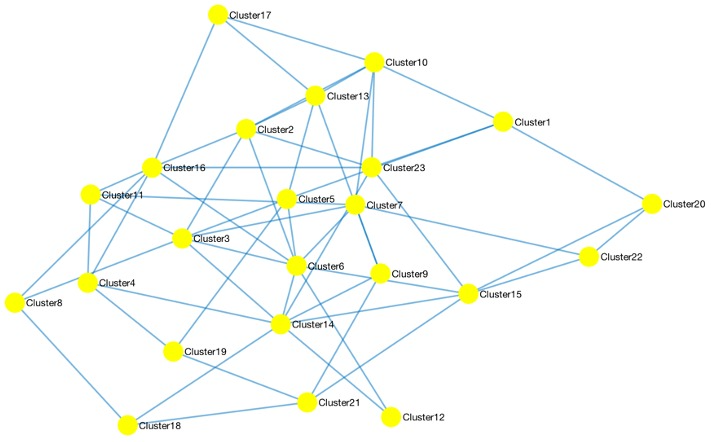
ATLAS uses a divide-and-conquer approach to generate large scale gene interaction networks for human transcriptomic data using experimental and external knowledge. Built upon our BNP framework, ATLAS uses probabilistic graph representations supplemented with known interaction information to identify its final network. Citation
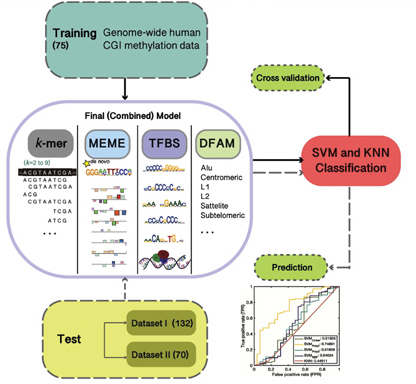
methylProp Predictor is an unbiased machine learning model that predicts the methylation propensity of CpG islands in the human genome using a combination of sequence-based biological features. Citation
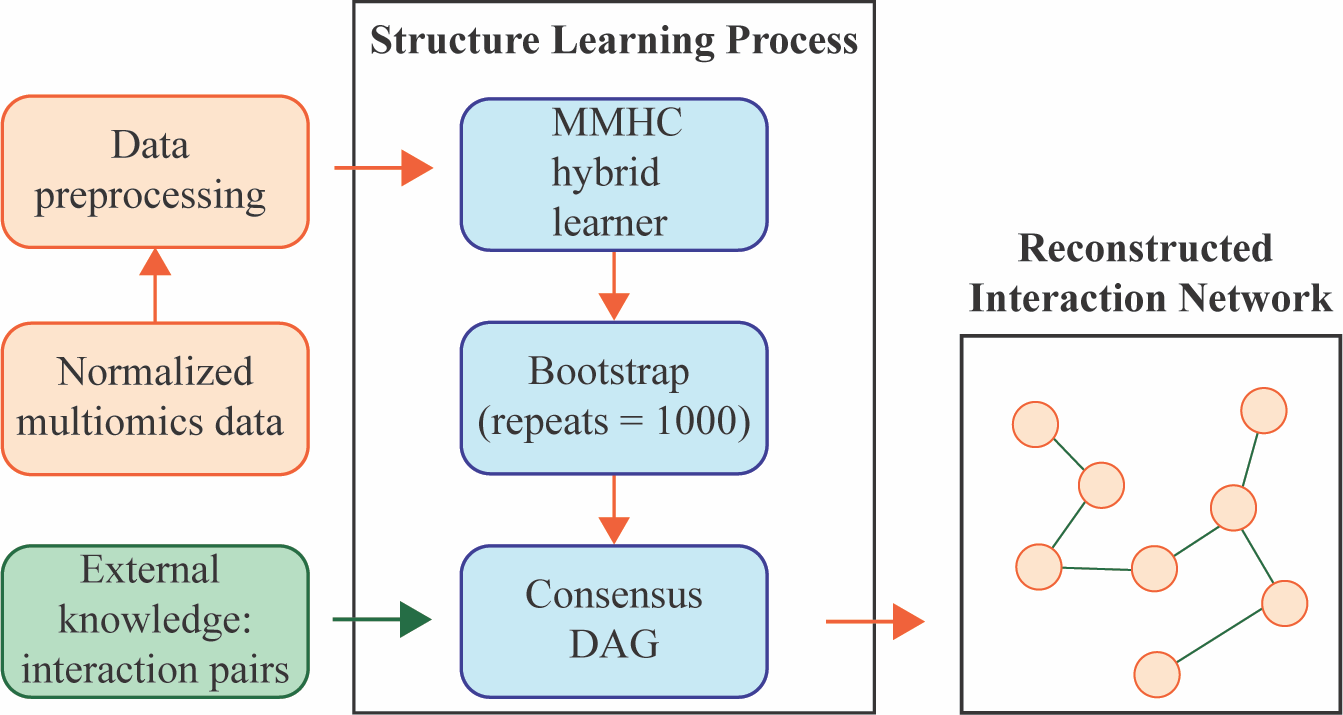
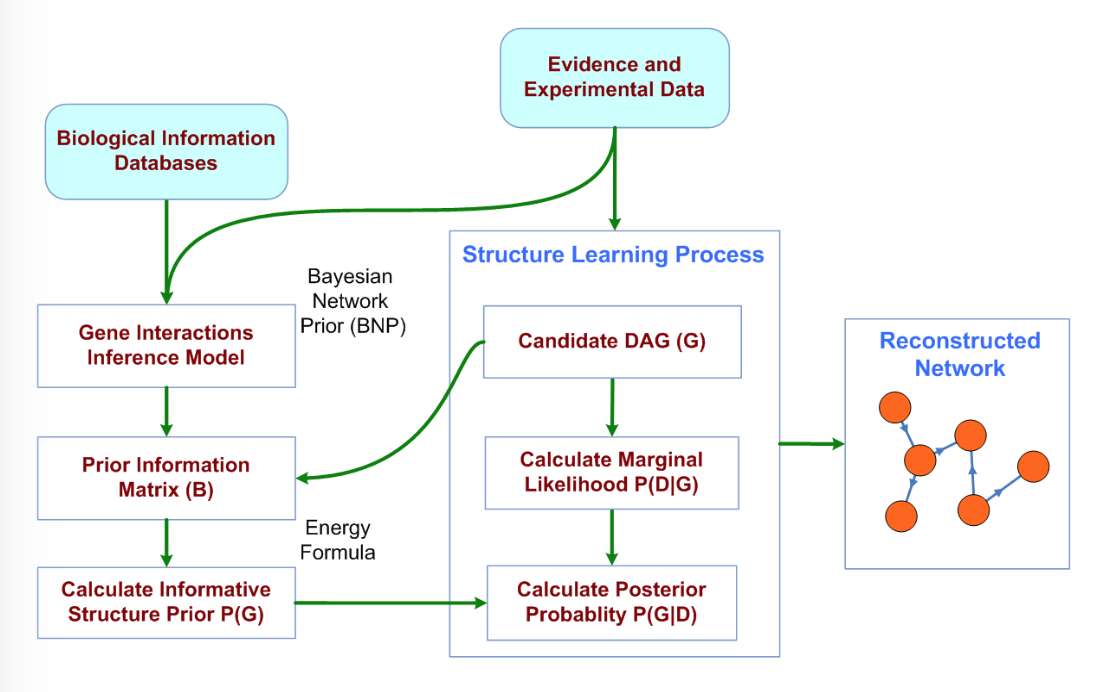
BNP (Bayesian Network Prior) provides a means to use external knowledge in building gene interaction networks using high-throughput biological data. BNP itself is a Bayesian Network that depicts the dependency structure between evidence types that imply an interaction event between two genes. Citation
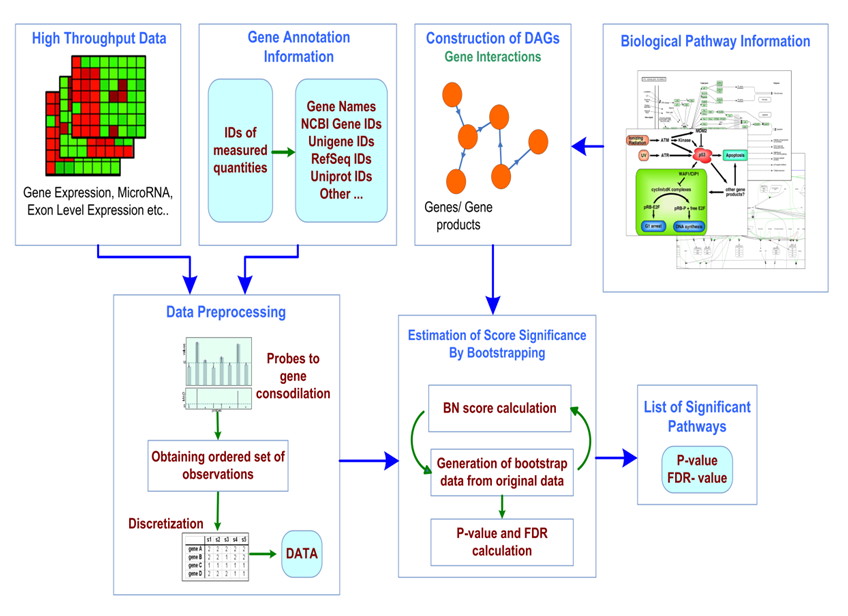
BPA (Bayesian Pathway Analysis) transcends popular enrichment analysis methods by considering the topology of a pathway when determining its association with given high-throughput biological data. Citation1 Citation2
OS Distance calculates the Otu-Sayood distance measure for a set of sequences. OS distance is an alignment-free approach that is based on an information-theoretic method to generate sequences. Citation
MineData (Coming soon) is a MATLAB-based user-friendly tool with enhanced GUI capabilities that provides mining multi-dimensional datasets using statistical, mathematical, and computational methods performing tasks like filtering, correlation, clustering, and classification analysis
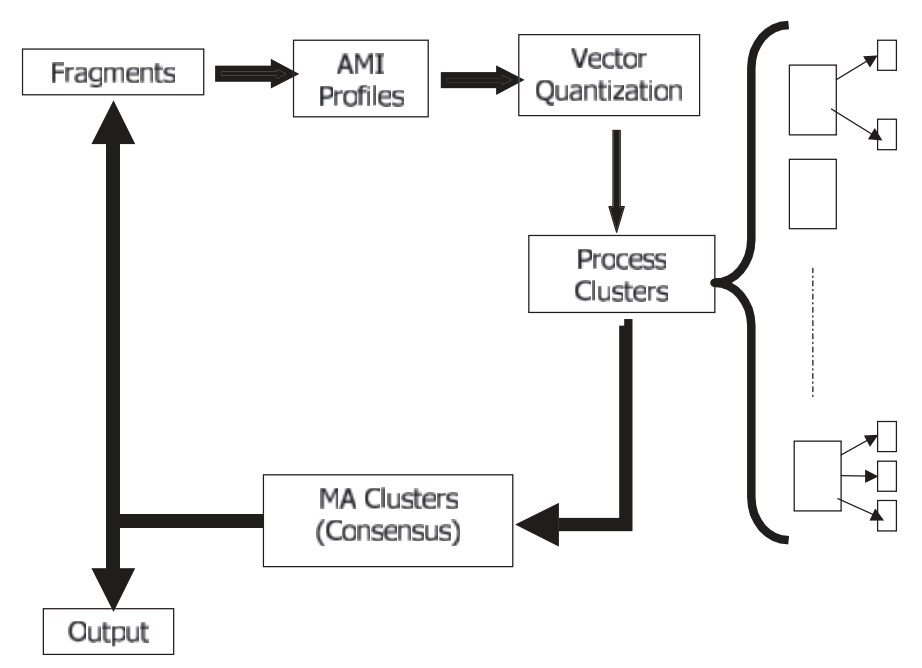
DAC-FS (A Divide And Conquer Approach to Fragment Assembly) is a computer program that calculates the contigs resulting from a shotgun sequencing data. DAC-FS simultaneously solves the overlap, layout, and consensus phases by iteratively clustering the fragments (or consensus sequences) with respect to their average mutual information (AMI) profiles and processing each cluster separately. Citation